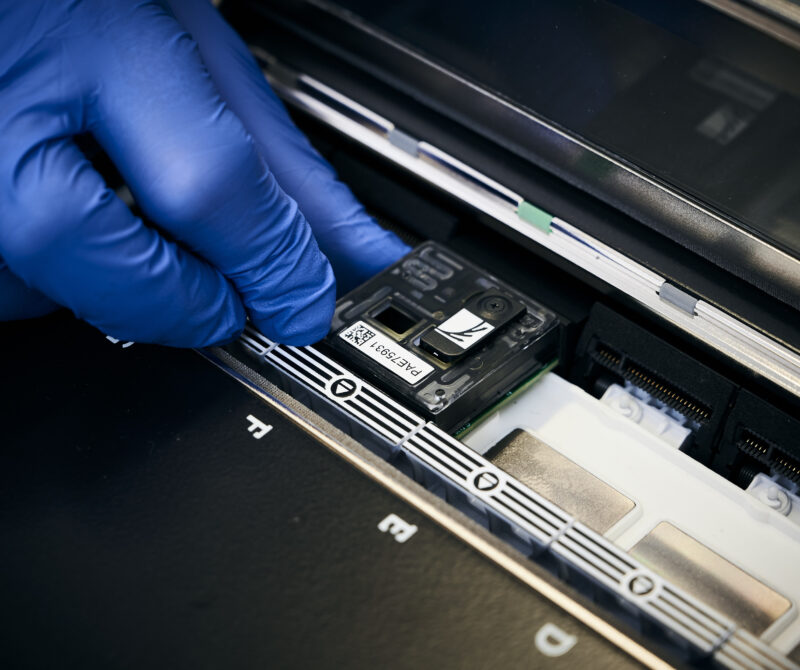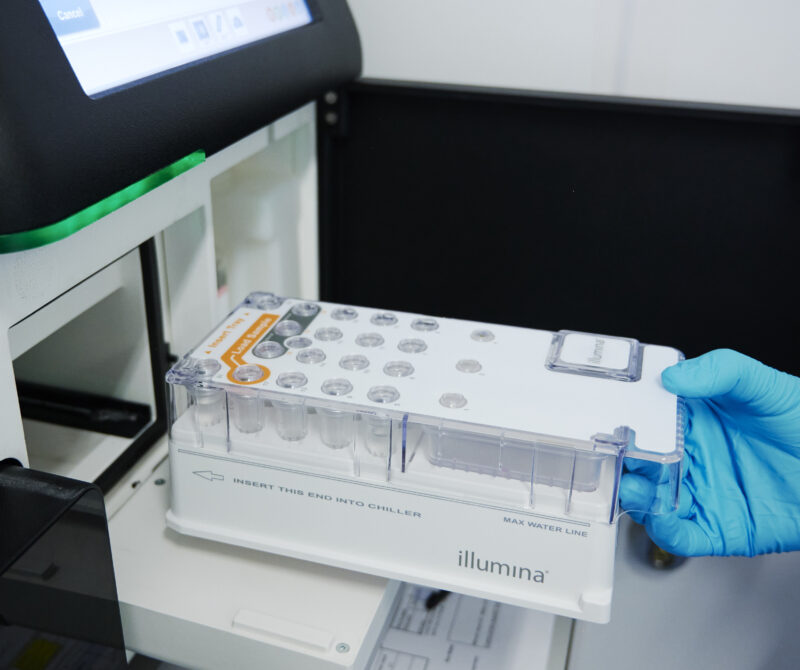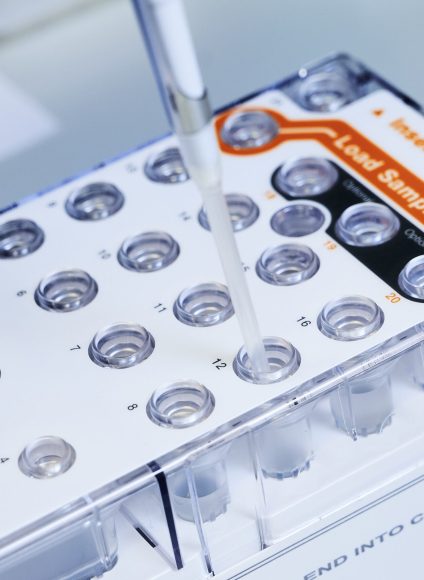
We are Edinburgh Genomics: The Genomics & Bioinformatics Core Facility at the University of Edinburgh
- We have established a global reputation for the provision of advanced genomics services
- We produce high-quality, high volume data for a diverse community of research collaborators and customers across academia, government and industry.
- Based at the University of Edinburgh's King's Buildings campus
Our Services
Sequencing

Short-read sequencing services using Illumina NovaSeq and MiSeq platforms, and long-read sequencing with the PacBio Revio and Oxford Nanopore PromethION 24 systems
Training

Introductory coding courses to advanced workshops on the latest bioinformatics tools delivered online and at the University of Edinburgh
Bioinformatics

Our dedicated team of research informaticians are skilled in all aspects of next generation genomics and genetic analyses
Sequencing

Short-read sequencing services using Illumina NovaSeq and MiSeq platforms, and long-read sequencing with the PacBio Revio and Oxford Nanopore PromethION 24 systems
Training

Introductory coding courses to advanced workshops on the latest bioinformatics tools delivered online and at the University of Edinburgh
Why Choose Us
Over 20 years experience
Versatile Sequencing Instrumentation
Recognised Facilities
Experienced training provider
Why Choose Us
Over 20 years experience
Versatile Sequencing Instrumentation
Recognised Facilities
Experienced training provider
22 +
1000 +
10 +


Testimonials
feedback from our collaborators and service users
“Really enjoyed the last few days of the “R for biologists course” of @edgenome. Thanks @nathcmedd for the great introduction! Can highly recommend to anyone who wants to have a first try with R”
“R for Biologists” Student, April 2022
“Really excellent. I learned so much and have never loved coding so much.”
“Introduction to Python for Biologists” Student, April 2017
“I really enjoyed the workshop and I thought it was extremely well organised and taught.”
“RNA-seq Data Analysis” Student, May 2017
“I thought it would be painful but it was actually fun!! #linux course. Thanks Tim & Nathan”
“Introduction to Linux for Genomics” Student, November 2021
“Having never used Linux before I came away with a working knowledge of the command-line and the confidence to use it.”
“Introduction to Linux for Genomics” Student, May 2016
“I also liked the fact that Martin spent a good amount of one on one time talking to people helping explain specific concepts and giving people advice on their personal academic projects.”
“Advanced Python for Biologists” Student, July 2016
“Loved it! I’m now itching to get my hands on my RNA-seq data and analyse it myself.”
“RNA-seq Data Analysis” Student, November 2014
“Great learning from someone with a biology background rather than computer science – meant we were on the same wavelength.”
“Introduction to Python for Biologists” Student, February 2016
“Just what I needed to start writing more complex code.”
“Advanced Python for Biologists” Student, August 2015
“Fantastic long reads bioinformatics workshop from @edgenome, superbly setup, good mix of theory and hands-on with @nanopore data. Also @urmi208 is a brilliant instructor, effortlessly handling a very mixed skills bunch of people – and all that, online, hats off!”
“Introduction to Long-read Bioinformatics” Student, May 2021
“Fantastic long reads bioinformatics workshop from @edgenome, superbly setup, good mix of theory and hands-on with @nanopore data. Also @urmi208 is a brilliant instructor, effortlessly handling a very mixed skills bunch of people – and all that, online, hats off!”
“Introduction to Long-read Bioinformatics” Student, May 2021
“Having never used Linux before I came away with a working knowledge of the command-line and the confidence to use it.”
“Introduction to Linux for Genomics” Student, May 2016
“I thought it would be painful but it was actually fun!! #linux course. Thanks Tim & Nathan”
“Introduction to Linux for Genomics” Student, November 2021
“I really enjoyed the workshop and I thought it was extremely well organised and taught.”
“RNA-seq Data Analysis” Student, May 2017
“Loved it! I’m now itching to get my hands on my RNA-seq data and analyse it myself.”
“RNA-seq Data Analysis” Student, November 2014
“I also liked the fact that Martin spent a good amount of one on one time talking to people helping explain specific concepts and giving people advice on their personal academic projects.”
“Advanced Python for Biologists” Student, July 2016


Testimonials
feedback from our collaborators and service users
“Great learning from someone with a biology background rather than computer science – meant we were on the same wavelength.”
“Introduction to Python for Biologists” Student, February 2016
“Really excellent. I learned so much and have never loved coding so much.”
“Introduction to Python for Biologists” Student, April 2017
“Really enjoyed the last few days of the “R for biologists course” of @edgenome. Thanks @nathcmedd for the great introduction! Can highly recommend to anyone who wants to have a first try with R”
“R for Biologists” Student, April 2022
“I really enjoyed the workshop and I thought it was extremely well organised and taught.”
“RNA-seq Data Analysis” Student, May 2017
“Having never used Linux before I came away with a working knowledge of the command-line and the confidence to use it.”
“Introduction to Linux for Genomics” Student, May 2016
“I also liked the fact that Martin spent a good amount of one on one time talking to people helping explain specific concepts and giving people advice on their personal academic projects.”
“Advanced Python for Biologists” Student, July 2016
“Loved it! I’m now itching to get my hands on my RNA-seq data and analyse it myself.”
“RNA-seq Data Analysis” Student, November 2014
“I thought it would be painful but it was actually fun!! #linux course. Thanks Tim & Nathan”
“Introduction to Linux for Genomics” Student, November 2021
“Thanks to brilliant @Nathcmedd for transforming me from completely clueless to vaguely competent in just 3 days! 100% recommend for anyone looking to get into coding!”
“R for Biologists” Student, August 2022
“Just what I needed to start writing more complex code.”
“Advanced Python for Biologists” Student, August 2015
“Fantastic long reads bioinformatics workshop from @edgenome, superbly setup, good mix of theory and hands-on with @nanopore data. Also @urmi208 is a brilliant instructor, effortlessly handling a very mixed skills bunch of people – and all that, online, hats off!”
“Introduction to Long-read Bioinformatics” Student, May 2021
“Having never used Linux before I came away with a working knowledge of the command-line and the confidence to use it.”
“Introduction to Linux for Genomics” Student, May 2016
“I thought it would be painful but it was actually fun!! #linux course. Thanks Tim & Nathan”
“Introduction to Linux for Genomics” Student, November 2021
“I really enjoyed the workshop and I thought it was extremely well organised and taught.”
“RNA-seq Data Analysis” Student, May 2017
“Loved it! I’m now itching to get my hands on my RNA-seq data and analyse it myself.”
“RNA-seq Data Analysis” Student, November 2014
“I also liked the fact that Martin spent a good amount of one on one time talking to people helping explain specific concepts and giving people advice on their personal academic projects.”
“Advanced Python for Biologists” Student, July 2016
Publications
1300 +
supported by our work

Upcoming Training Courses
100 +
Upcoming Events
News
65th NextgenBUG Meeting in Edinburgh, May 30th
Edinburgh Genomics acquires a PacBio Revio System
🧬 Last week, we ran a #DNA Library Prep for Short Read #Sequencing workshop! 🧪
The course was fully booked with 12 keen participants, and we were delighted to have reps from @NEBiolabs join us to support and share insights with attendees.🦋
Thanks to everyone who took part!👏
Last week, we hosted a fantastic seminar with @illumina on single-cell analysis! Great discussions, insightful questions, and plenty of inspiration for future research.
Thanks to everyone who joined, and especially to Laura and Pauline from @illumina !
#Genomics #SingleCell
Do you have an enquiry, or would you like to get in touch?
Do you have an enquiry, or would you like to get in touch?
Natural Environment Research Council

Biotechnology and Biological Sciences Research Council

Medical Research Council

Scottish Universities Life Sciences Alliance